 DNA origami is the folding of DNA to create non-arbitrary two- and three-dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material, through design of its base sequences. DNA is a well-understood material that is suitable for creating scaffolds that hold other molecules in place or to create structures all on its own.
DNA origami is the folding of DNA to create non-arbitrary two- and three-dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material, through design of its base sequences. DNA is a well-understood material that is suitable for creating scaffolds that hold other molecules in place or to create structures all on its own.

Build a DNA origami structure
We begin the design of DNA origami using cadDNAno software. cadDNAno simplifies and enhances the process of designing three-dimensional DNA origami nanostructures. Through its user-friendly 2D and 3D interfaces it accelerates the creation of arbitrary designs. The embedded rules within cadDNAno paired with the finite element analysis performed by CanDo, provide relative certainty of the stability of the structures. 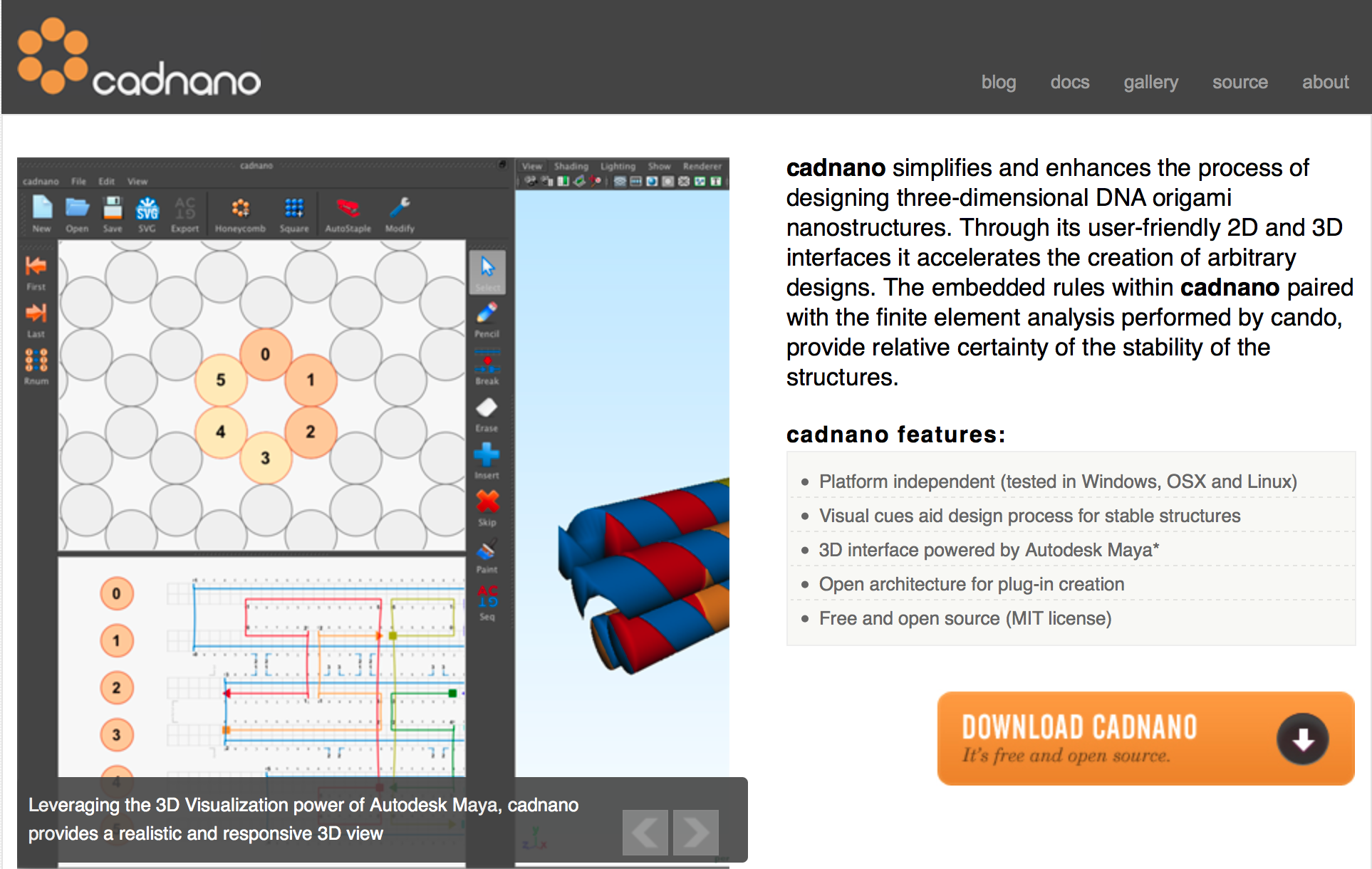
1. Build flattened Rothemund rectangle in cadDNAno

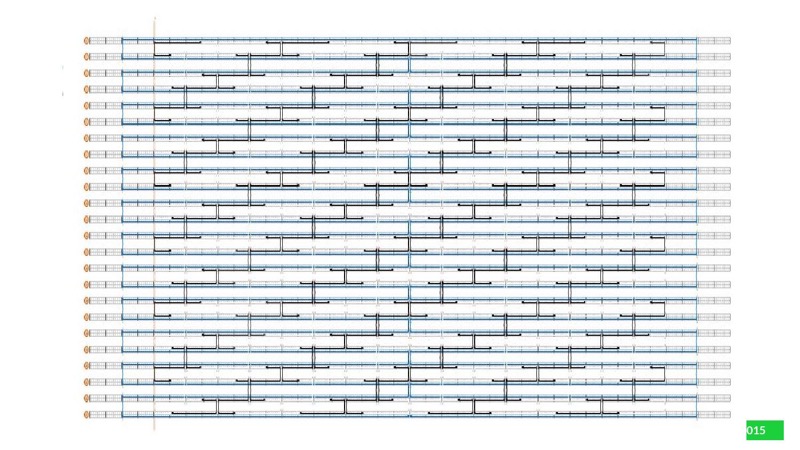
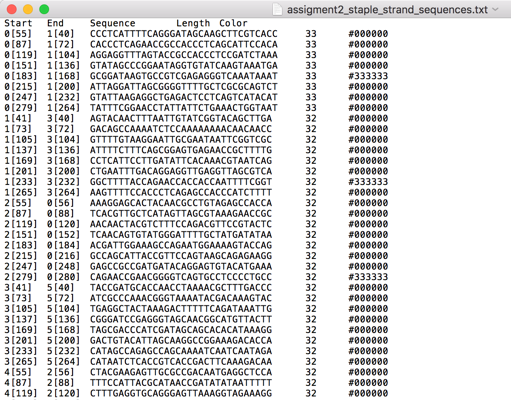
2. Generate staple strand sequences in caDNAno
3. Color staple strands to create a black-and-white pattern in caDNAno
JSON file output for the black-and-white pattern in cadDNAno:
4. Generate ASCII file black-and-white image

5. Generate list of wells to pipet to generate black-and-white pattern
By parsing the black-and-white ASCII pattern, we selected the appropriate empty and dumbbell staple DNA oligos from the respective 96-well plates. You can see the compiled pipeting instructions in Fig. 5. (The list is given in a 2D format, but it is essentially linear; you pipet everything and mix!

Generate caDNAno-format json file encoding 24 strand flat rectangle from a Python script
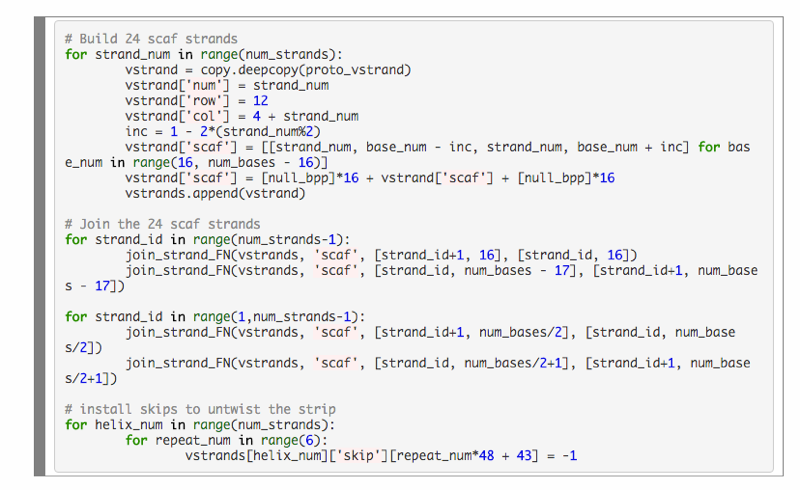
Experimental Protocol
![How to grow [almost] anything class Viirj Kan](img/logo.png)