
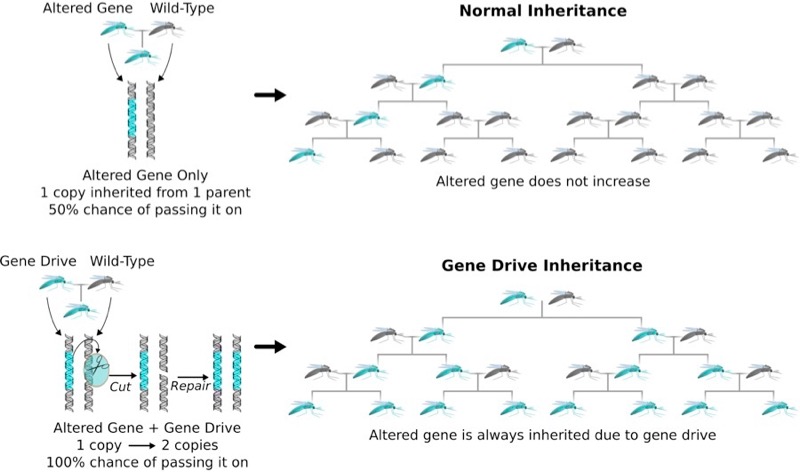
Design a basic but evolutionarily stable gene drive that should function in your organism
The goal is to design a proof-of-principle experiment that will determine whether CRISPR gene drives can function efficiently in the target organism. The drive system should not cause population suppression, carry any 'cargo' genes, or change the sequence of any protein produced by the organism. It should only spread itself. An example is this proposal to build a gene drive capable to eradicating schistosomiasisIf the genome sequence for your organism isn't available or is hard to work with in any way, use the nematode worm Caenorhabditis elegans instead – we may use your design for a hands-on experiment in next year's class.
Identify a problem that could be addressed using a CRISPR gene drive

However, many things could go wrong without careful design as the Gypsy moth uses its olfactory receptors also for mating, which means that altering the target odorant repertoire could putatively render them unable to detect their mates. In that case, the gene drive would not be effective, but fortunately the effect will not be catastrophic (suppress the population) as by inhibiting mating we are essentially stripping the ability of the drive to be passed to the next generation.
1) Identify a gene that is important for fitness in your target organism. Why must gene drives target genes important for fitness? A literature search may be required. You can use NCBI for information on genes if you wish, though Wormbase is generally superior and easier to use.
The gene we choose to target is the OrCo gene (sequence shown below from NCBI), which stands for olfactory receptor co-receptor. Knockout of Ors has revealed that OrCo proteins are required for insect olfaction. The gene must be important for fitness because if the gene makes the organism less competitive, then the organism will most likely die before propagating the gene to next generation.
2) Find CRISPR target sites in the 3' end of the gene using an online tool. For C. elegans and many other organisms, a good user-friendly tool is GT-Scan. Recommended parameters: set the high-specificity mismatch limit to 3 and leave all other settings at default values. More exotic species require less user-friendly software. For bonus points, use sgRNAcas9, which allows you to analyze any downloaded genome sequence in fasta format. Warning: it's not particularly user-friendly. List the relevant criteria for inclusion of each target sequence (e.g. potential off-targets etc) as provided by the program you used.
The genome for lymantria dispar is not sequencedWe will answer this portion of the homework as a hypothetical. If the genome was hypothetically available, we would design the CRISPR target sites based on the sgRNAcas9 software (since this species is not available on GT-scan) -- we run the OrCo gene against the gypsy moth genome and design a GUIDE RNA that does not exist anywhere else in the moth genome. This ensures specificity. Also, it is important to include the target sequence at the 3' end of the OrCo gene.
Alternatively, we started a new search beginning with a common pest whose genome is sequenced. We found the Fall Armyworm (Spodoptera frugiperda), seen in a paper. However, as the paper explains, the genome has not been annotated, at least not for olfactory receptors. What we did was run a BLAST search of common known olfactory receptors against the sequenced armyworm. We found around 10 olfactory genes with around 95% match to the armyworm genome. From this data we can then run a GT-scan and design the proper guide-RNA and point of insertion near the 3' end.
3) Would it be possible for you to safely build this gene drive in your current laboratory? Which confinement strategies and safeguards would you use? See Akbari et al. and especially here for recommended confinement strategies.
It is definitely not safe for us to build the gene drive -as is- in our current laboratory as pests like Gypsy moth can be found all over the US and can be transferred accidentally on crop transportation. We would use both extrinsic and intrinsic confinement methods. As moths can fly, we would start with a barrier confinement made by strong air curtains on the doors of the laboratory and mandatory inspection of the lab clothing prior to exit for moths. The moths will be kept in multiple nested containers that are only ever opened when their occupants have been anesthetized. At the same time, we can use molecular confinement strategies.4) Extra credit: Generate a version of the target gene that will be carried with your gene drive. Recode the target gene so that it encodes the same protein, but does not have the sequences you are targeting. Should you change only those codons, most, or all of them?
Envision an Open Discussion Forum for Gene Drives
What features would you want to see in an online discussion platform devoted to guiding the development of gene drives? Assume that the researchers involved in the project are interested in soliciting public feedback before and during experiments so that they can better identify problems and redesign the technology. Please give specific examples of already-existing elements – if you want a discussion forum, should it be more like reddit, Quora, or something else? What should moderation be like? What would it take to get you to regularly participate in such a community?
Media outlets are a double-edged sword: they are some of the primary venues to receive news (including science news) but the portrayal and delivery can be sensationalized, polemic, biased. In the case of new technology as controversial as CRISPR gene drives, it can potentially spark a lot of emotive responses on the part of the public in regards to what this technology can impact. Scientists don't always take the time for science communication, engaging with the public on their research, which is especially important if their research has potential downfalls. However, some benefits could come from a direct bridge of communication from scientists to the public, cutting out the middleman which is the media. In addition, the public should not be expected to comprehend heavy scientific jargon that is present in published articles and textbooks, so a scientist who is able to translate esoteric knowledge into terms of the lay-public would be especially beneficial. Finally, there is the issue of making people care. Why would people care about gene drives, enough that they actively participate in a public discussion? Is it enough that the technology is just THAT controversial that immediately we can expect the public to open their eyes and pay full attention? This is almost a question for biotechnology in general -- how to reveal the profound impacts so that people start to care. So then, we should start discussions with the applications:
![How to grow [almost] anything class Viirj Kan](img/logo.png)